Our research focuses on the development of machine learning approaches to determine the structure of macromolecular complexes of general biomedical interest using single-particle cryo-electron microscopy, cryo-electron tomography, and sub-volume averaging. Some of our targets include glycoproteins of enveloped viruses like HIV, Influenza and Ebola, transporters and channels involved in signaling and metabolism, GPCRs, DNA-targeting CRISPR-Cas surveillance complexes, and targets for cancer drugs.
We are also interested more broadly in deep learning and artificial intelligence, computer vision, biomedical imaging, and high-performance computing.
Featured Research
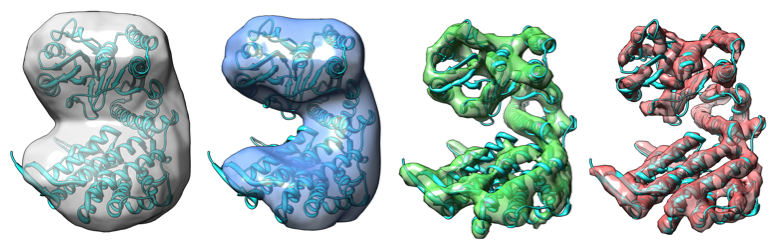
Improvements in the detective quantum efficiency of direct electron detectors combined with their ability to operate in movie mode have created unique opportunities for the development of computational strategies in single-particle cryo-EM capable of achieving unprecedented resolutions. To harness the time-resolved information contained in movie frames, we developed methods to account for local variations in defocus and beam-induced drift, and implemented a data-driven dose compensation scheme to more efficiently extract the high-resolution information recorded during the electron exposure, enabling for the first time the visualization of atomic resolution features in density maps obtained using single-particle Cryo-EM.

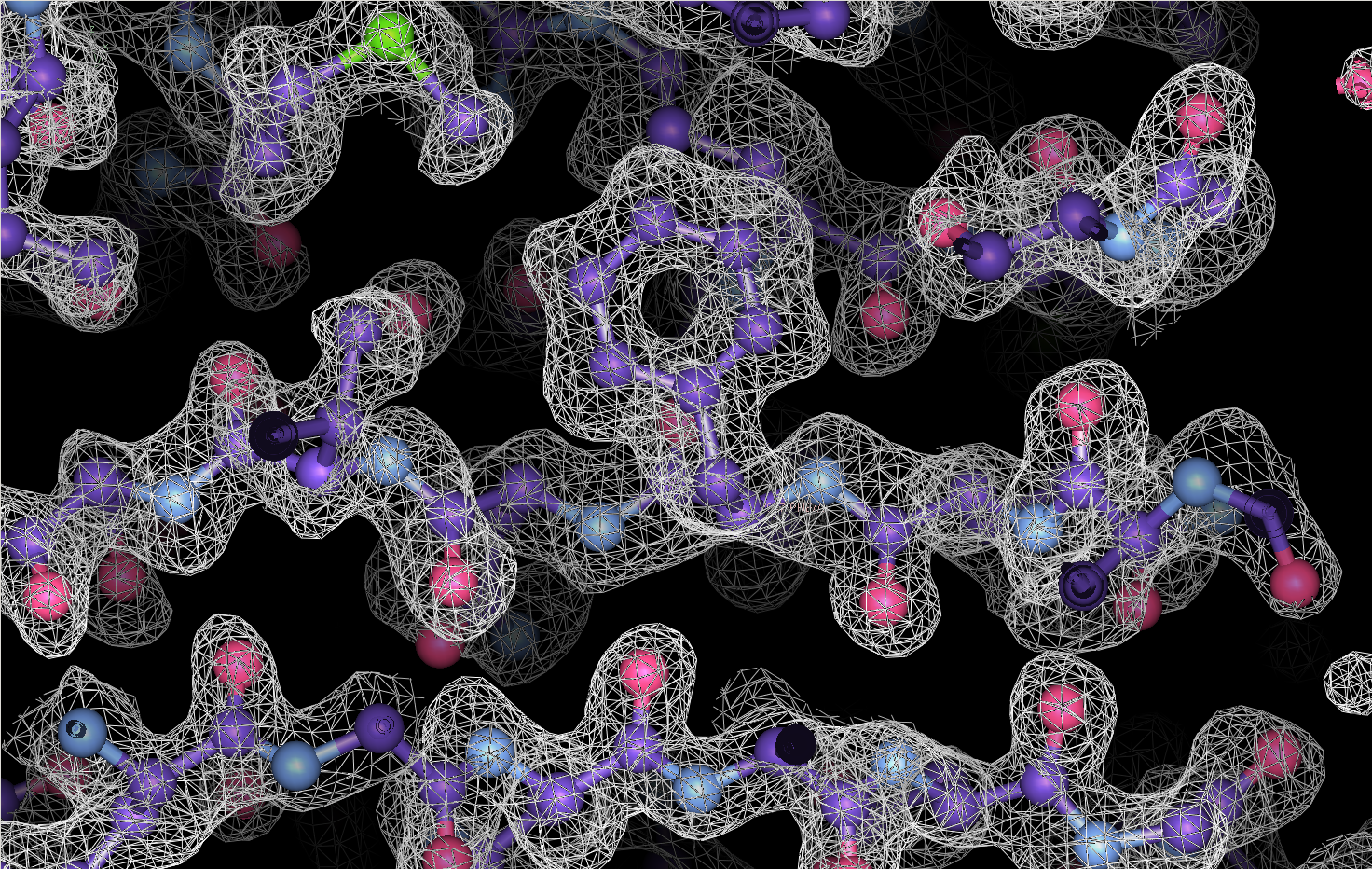
Cryo-EM is a powerful technique for structure determination of protein complexes by averaging information from individual molecular images. Tomographic data collection schemes provide powerful constraints that can be used to more accurately determine molecular orientations necessary for 3D reconstruction. Constrained Single-Particle Tomography is a general strategy for 3D structure determination in cryo-EM that effectively uses images recorded in tilt series to extract high-resolution information and correct for the contrast transfer function of the electron microscope. By incorporating geometric constraints into the refinement of image orientations, we demonstrate that protein structures can be determined at significantly higher resolutions than those obtained using conventional sub-tomogram averaging.