Skip to content
-
-
- Cryo-EM, Methods, Publications
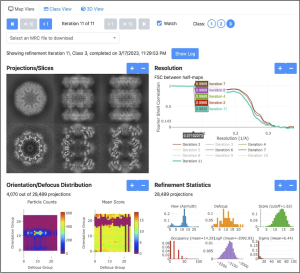
- nextPYP: a comprehensive and scalable platform for characterizing protein variability in situ using single-particle cryo-ET Single-particle cryo-electron tomography (SP-CET) is an emerging technique capable of determining the structure of proteins imaged within the native context of cells at molecular resolution. While high-throughput techniques for sample preparation and tilt-series acquisition are beginning to provide sufficient data to allow structural studies…
-
-
- Cryo-EM, Methods, Publications
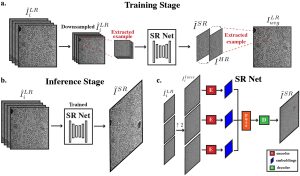
- Multiple-image super-resolution of cryo-electron micrographs based on deep internal learning Single-particle cryo-electron microscopy (cryo-EM) is a powerful imaging modality capable of visualizing proteins and macromolecular complexes at near-atomic resolution. The low electron-doses used to prevent radiation damage to the biological samples, however, result in images where the power of the noise is 100 times greater than the power of…
-
-
- Publications, Structures
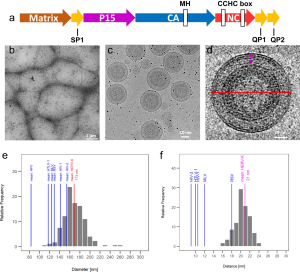
- Molecular architecture and conservation of an immature human endogenous retrovirus The human endogenous retrovirus K (HERV-K) is the most recently acquired endogenous retrovirus in the human genome and is activated and expressed in many cancers and amyotrophic lateral sclerosis. We present the immature HERV-K capsid structure at 3.2 Å resolution determined from native virus-like particles using cryo-electron tomography and subtomogram…
-
-
- HIV, Publications, Structures
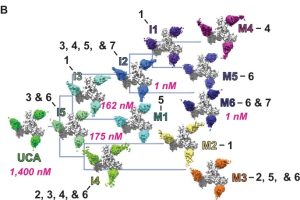
- Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage Antibody affinity maturation enables adaptive immune responses to a wide range of pathogens. In some individuals broadly neutralizing antibodies develop to recognize rapidly mutating pathogens with extensive sequence diversity. Vaccine design for pathogens such as HIV-1 and influenza has therefore focused on recapitulating the natural…
-
-
- Publications, Structures
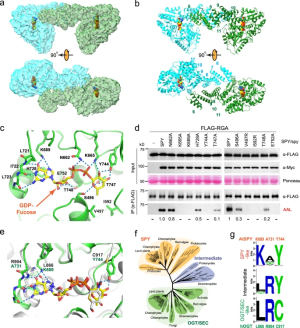
- Structure and dynamics of the Arabidopsis O-fucosyltransferase SPINDLY SPINDLY (SPY) in Arabidopsis thaliana is a novel nucleocytoplasmic protein O-fucosyltransferase (POFUT), which regulates diverse developmental processes. Sequence analysis indicates that SPY is distinct from ER-localized POFUTs and contains N-terminal tetratricopeptide repeats (TPRs) and a C-terminal catalytic domain resembling the O-linked-N-acetylglucosamine (GlcNAc) transferases (OGTs). However, the structural feature that determines the…
-
-
- Cryo-EM, Methods, Publications
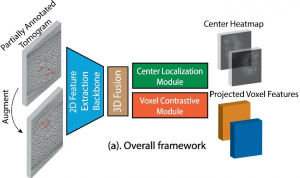
- Accurate Detection of Proteins in Cryo-Electron Tomograms from Sparse Labels Cryo-electron tomography (CET) combined with sub-volume averaging (SVA), is currently the only imaging technique capable of determining the structure of proteins imaged inside cells at molecular resolution. To obtain high-resolution reconstructions, sub-volumes containing randomly distributed copies of the protein of interest need be identified, extracted and subjected to SVA,…
-
-
- Cryo-EM, Methods, Publications
- High-resolution structure determination using high-throughput electron cryo-tomography The low throughput characteristic of tomographic data acquisition combined with the complex data-analysis pipeline that is required to obtain high-resolution maps, has limited the applicability of this technique to favorable samples or to resolutions that are too low to provide useful mechanistic information. Recently, beam image-shift electron cryo-tomography (BISECT), a strategy to…
-
-
- Publications, Ribosome, Structures
- Redox-sensitive E2 Rad6 controls cellular response to oxidative stress via K63-linked ubiquitination of ribosomes In this study, we set out to investigate the key role of Rad6 in regulating cellular response to stress in budding yeast as part of the RTU. Rad6 is small (20 kDa), highly conserved, and a multifunctional E2 involved in DNA repair and in the…